A recent study titled “Structural basis of nucleosome recognition and modification by MLL methyltransferases” published online in Nature on September 04 from the research group of Dr. Jing Huang reported the cryo-electron microscopy (cryo-EM) structures of human MLL methyltransferases in complex with histone H2B-monoubiquitinated nucleosomes or unmodified nucleosomes. MLL family methyltransferases implement histone H3K4 methylation and play critical roles in transcription regulation of hematopoiesis, adipogenesis, and development. Mechanistic elucidation for activity regulation of MLL family methyltransferases is important for understanding their multifaceted cellular functions as well as their pathogenic roles in MLL-related diseases. This work provides an unprecedented structural framework for the activation mechanism of MLL, which for the first time reveals that the nucleosome substrate stimulates the enzymatic activities of MLL proteins through multiple specific interactions with MLL and its regulatory factors (WDR5, RBBP5, ASH2L, and DPY30, abbreviated as WRAD). Prior to this study, it has long been assumed that the protruding histone H3 tail is readily accessible to the MLL proteins, and the structural characterization of MLL complexes has been limited to the catalysis of synthetic histone H3 peptides. Cryo-EM structures of human MLL1 and MLL3 complexes associated with H2B-monoubiquitinated nucleosomes clearly demonstrate the pivotal roles of nucleosome-binding in the activity regulation of MLL family methyltransferases. This means that deposition of H3K4-methyl marks could be controlled by higher-order chromatin structures as well as pre-existing histone marks or chromatin-binding factors to achieve a complex and integrated regulation of this molecular process in cells. This finding also has great implications in the mechanistic investigations of other histone tail modifiers, suggesting that nucleosome core regions might be involved in the activity regulation of other posttranslational modifications of histone tails. The activation mechanism of MLL by nucleosome substrates also explains how monoubiquitination of H2B lysine 120 (H2BK120ub1) stimulates H3K4 methylation. Cryo-EM structures of MLL1/3-ubNCP complexes reveal that the H2BK120-conjugated ubiquitin directly binds to the WD40 domain of RBBP5, orientating the MLL-nucleosome interactions. Six members of human MLL family (MLL1-MLL4, SET1A, and SET1B) have distinct cellular functions that are closely related to their specific methyltransferase activities. Cryo-EM structures of MLL1 (di- and tri-methyltransferase) and MLL3 (mono-methyltransferase) complexes reveal a critical structural mechanism for the activity difference between the two MLL complexes. In the MLL1 complex, an activation segment of MLL1 (MLL1AS), which resembles that of other SET-domain-containing methyltransferases for optimal activities of MLL1, was identified. The conformation of MLL1AS is stabilized by the WDR5-MLL1-RBBP5 tri-subunit interactions. However, in the MLL3 complex, lack of an MLL1AS counterpart at the WDR5-MLL3-RBBP5 tri-subunit interface leads to direct contact between WDR5 and MLL3SET, which inhibits the intrinsic activities of MLL3. WDR5 plays diverse roles in the assembly and activity regulation of MLL family complexes. It is also targeted for cancer therapy development. This work provides a fundamental mechanistic explanation for the roles of WDR5 in finetuning the enzymatic activities of MLL members for various cellular functions. In summary, this work reveals the structural mechanisms of nucleosome recognition and activity specificity of human MLL family methyltransferase, and brings up the concept of nucleosomal regulation on histone tail modification. These findings will promote the mechanistic understanding of cellular functions of MLL proteins as well as the therapy development against MLL-related diseases. Download link: https://www.nature.com/articles/s41586-019-1528-1
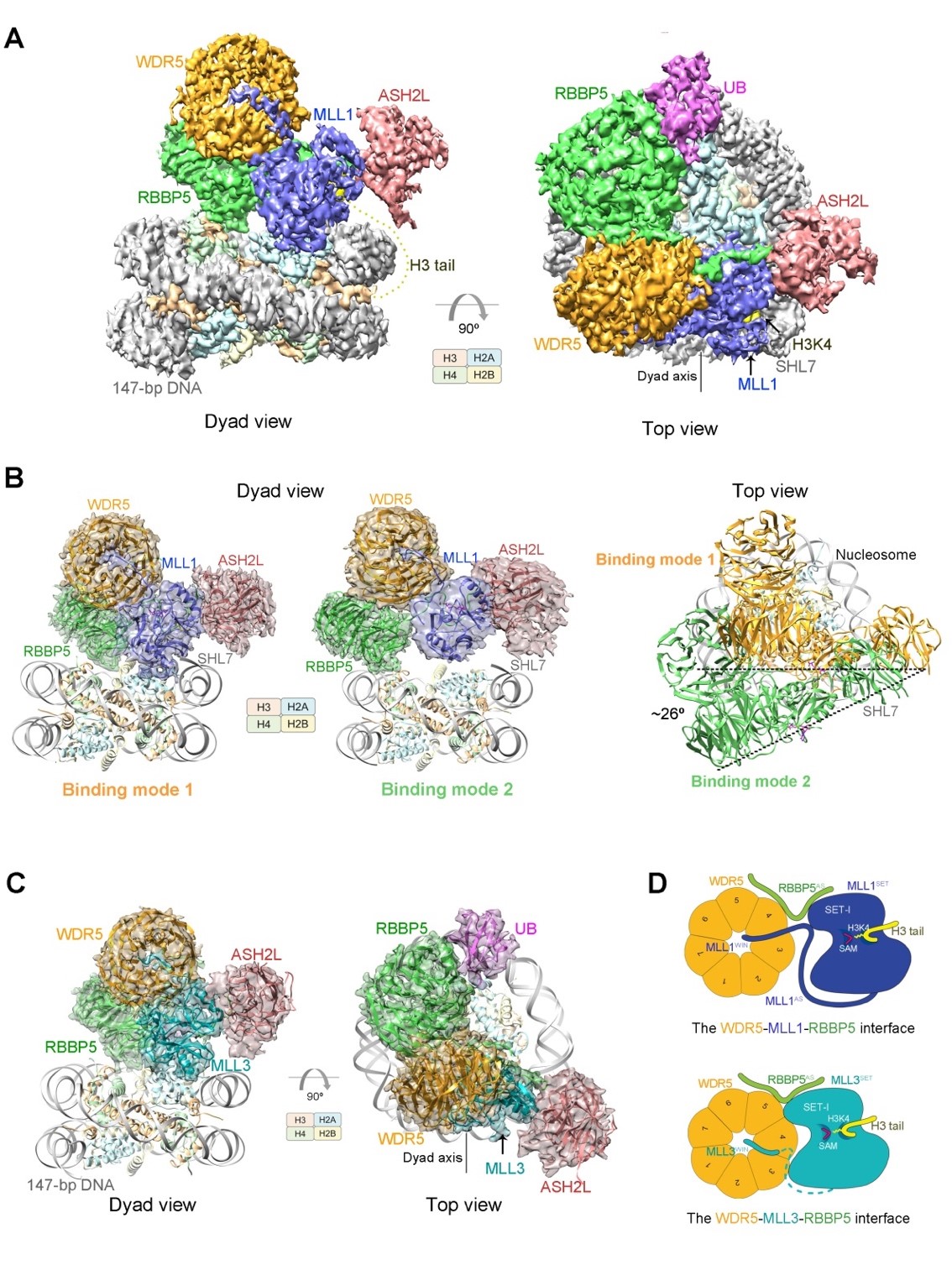
Fig.1 Cryo-EM structures of human MLL1 complex and MLL3 complex associated with unmodified or ubiquitinated nucleosomes. (A) Cryo-EM density map of human MLL1-ubNCP complex shown from two orthogonal views. (B) Cryo-EM density maps and atomic models of the two binding-modes of human MLL1-NCP complex in the dyad view of nucleosome. The right panel is the superposition of the two MLL1-NCP structures, showing the rotation of the MLL1 complex on nucleosome surface between the two binding modes. (C) Cryo-EM density map and atomic model of human MLL3-ubNCP complex shown from two orthogonal views. (D) A working model shows the different structural organizations at the WDR5-MLL1/3-RBBP5 interfaces that result in the activity difference in MLL1 and MLL3 complexes.
|